ANCOVA code in Python
This article explains how to perform the one-way ANCOVA in Python. You can refer to this article to know more about ANCOVA, when to use ANCOVA, assumptions, and how to interpret the ANCOVA results.
One-way (one factor) ANCOVA in Python
Load the dataset
import pandas as pd
df=pd.read_csv("https://reneshbedre.github.io/assets/posts/ancova/ancova_data.csv")
df.head(2)
genotype height yield
0 A 10.0 20.0
1 A 11.5 22.0
Summary statistics and visualization of dataset
Get summary statistics based on dependent variable and covariate,
from dfply import *
# summary statistics for dependent variable yield
df >> group_by(X.genotype) >> summarize(n=X['yield'].count(), mean=X['yield'].mean(), std=X['yield'].std())
# output
genotype n mean std
0 A 10 25.17 2.577704
1 B 10 35.45 2.431849
2 C 10 17.73 2.039635
# summary statistics for covariate height
df >> group_by(X.genotype) >> summarize(n=X['height'].count(), mean=X['height'].mean(), std=X['height'].std())
# output
genotype n mean std
0 A 10 13.790 2.224835
1 B 10 16.756 1.963960
2 C 10 9.600 1.840290
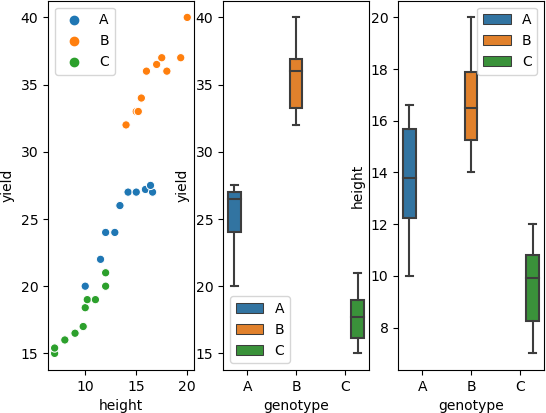
Visualize dataset,
import seaborn as sns
import matplotlib.pyplot as plt
fig, axs = plt.subplots(ncols=3)
sns.scatterplot(data=df, x="height", y="yield", hue=df.genotype.tolist(), ax=axs[0])
sns.boxplot(data=df, x="genotype", y="yield", hue=df.genotype.tolist(), ax=axs[1])
sns.boxplot(data=df, x="genotype", y="height", hue=df.genotype.tolist(), ax=axs[2])
plt.show()
perform one-way ANCOVA
from pingouin import ancova
ancova(data=df, dv='yield', covar='height', between='genotype')
# output
Source SS DF F p-unc np2
0 genotype 193.232458 2 141.352709 1.072947e-14 0.915777
1 height 132.695696 1 194.138152 1.429323e-13 0.881892
2 Residual 17.771304 26 NaN NaN NaN
If you have any questions, comments or recommendations, please email me at reneshbe@gmail.com
This work is licensed under a Creative Commons Attribution 4.0 International License

